
真核生物剪接体可通过选择性拼接事件将不同的外显子拼接到一起,以组成多样的转录本,进而形成功能多样的蛋白质。环状RNA是一类由mRNA前体经反向拼接形成的共价闭合环状非编码RNA,在响应植物胁迫和开花调控上具有重要功能。R-loop是一种三链核酸结构,包括RNA和它的模板DNA缠绕形成的杂合双链和一条非模板DNA单链,拟南芥中的研究表明R-loop结构广泛存在。但环状RNA是否通过R-loop结构广泛地调控选择性拼接事件依然不清楚。JIPB近日在线发表了福建农林大学海峡联合研究院林学中心顾连峰课题组题为“Genome-wide profiling of circular RNAs, alternative splicing, and R-loops in stem-differentiating xylem of Populus trichocarpa”(https://doi.org/10.1111/jipb.13081)的研究论文。以毛果杨次生木质部为研究材料,通过杂合链免疫共沉淀及高通量测序技术(DRIP-seq)、环状RNA测序和PacBio异构体测序等组学技术,分别检测了毛果杨次生木质部中的R-loop结构、环状RNA和选择性拼接事件,组学关联分析发现环状RNA明显在R-loop结构和选择性拼接事件区域富集。过表达和次生生长相关的circ-IRX7,发现过表达样本可以检测到R-loop结构增加,并影响了宿主基因的选择性拼接,而过表达没有R-loop结构的circ-GUX1则未能观察到宿主基因选择性拼接的变化。这为后续研究环状RNA的拼接调控功能提供了初步的线索。
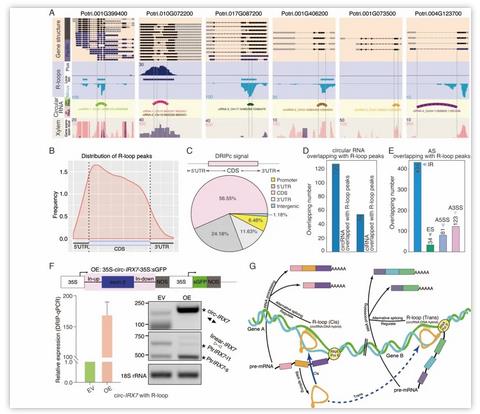
环状RNA通过R-loop结构调控宿主基因的选择性拼接
顾连峰课题组近年来在林木环状RNA研究方面取得进展:发现DNA甲基化在环状RNA侧翼内含子区域富集 (Zhang et al., 2020);环状RNA和线形转录本一样,也受到转录后调控 (Wang et al., 2019a)和N6-甲基腺嘌呤修饰的调控 (Wang et al., 2020);开发了环状RNA的可视化流程和RNA表观修饰的检测方法 (Gao et al., 2018; Gao et al., 2021);发现环状RNA可调控宿主基因的表达 (Wang et al., 2019b);这篇论文进一步揭示部分环状RNA可通过和DNA形成R-loop结构调控宿主基因的选择性拼接。福建农林大学林学院博士生刘旭庆和高宇帮为该论文的第一作者,顾连峰教授为通讯作者。林学中心研究生廖家凯、陈凯和林学院本科生苗苗等也参与该项工作。该研究得到了国家重点研发项目(2016YFD0600106)的支持。
1.Gao, Y., Liu, X., Wu, B., Wang, H., Xi, F., Kohnen, M.V., Reddy, A.S. and Gu, L. (2021). Quantitative profiling of N6-methyladenosine at single-base resolution in stem-differentiating xylem of Populus trichocarpa using Nanopore direct RNA sequencing. Genome Biol. 22: 1-17.2. Gao, Y., Wang, H., Zhang, H., Wang, Y., Chen, J., and Gu, L. (2018). PRAPI: Post-transcriptional regulation analysis pipeline for Iso-Seq. Bioinformatics 34: 1580-1582.3. Wang, H., Wang, H., Zhang, H., Liu, S., Wang, Y., Gao, Y., Xi, F., Zhao, L., Liu, B., Reddy, A.S.N., Lin, C., and Gu, L. (2019a). The interplay between microRNA and alternative splicing of linear and circular RNAs in eleven plant species. Bioinformatics 35: 3119-3126.4. Wang, Y., Wang, H., Xi, F., Wang, H., Han, X., Wei, W., Zhang, H., Zhang, Q., Zheng, Y., Zhu, Q., Kohnen, M.V., Reddy, A.S.N., and Gu, L. (2020). Profiling of circular RNA N(6) -methyladenosine in moso bamboo (Phyllostachys edulis) using nanopore-based direct RNA sequencing. J. Integr. Plant Biol. 62: 1823-1838.5. Wang, Y., Gao, Y., Zhang, H., Wang, H., Liu, X., Xu, X., Zhang, Z., Kohnen, M.V., Hu, K., Wang, H., Xi, F., Zhao, L., Lin, C., and Gu, L. (2019b). Genome-wide profiling of circular rnas in the rapidly growing shoots of moso bamboo (Phyllostachys edulis). Plant Cell Physiol. 60: 1354-1373.6. Zhang, Z., Wang, H., Wang, Y., Xi, F., Wang, H., Kohnen, M.V., Gao, P., Wei, W., Chen, K., Liu, X., Gao, Y., Han, X., Hu, K., Zhang, H., Zhu, Q., Zheng, Y., Liu, B., Ahmad, A., Hsu, Y.-H., Jacobsen, S.E., and Gu, L. (2021) Whole genome characterization of chronological age-associated changes in methylome and circular RNAs in moso bamboo (Phyllostachys edulis) from vegetative to floral growth. Plant J. DOI: 10.1111/tpj.15174.











