 | 主任(PI): 朱强(Qiang Zhu, PhD) |
团队成员(Members):王文佳(Wenjia Wang),唐晓珊(Xiaoshan Tang),叶善汶(Shanwen Ye),蔡昌杨(Changyang Cai),朱彩萍(Caiping Zhu),张莉(Li Zhang)
PI简介(PI Introduction):
朱强,教授,博士生导师。2004年于鲁东大学取得生物学学士学位;2009年获得中国科学院上海植物生理生态研究所遗传学博士学位。2010-2015年分别在比利时根特大学(Ghent University)和奥地利科学技术研究院(IST Austria)进行博士后研究,主要从事的研究包括植物抗逆分子机制;RNA编缉因子在植物生长发育和抗逆方面的作用;双子叶植物顶端弯钩的发育机理等。目前主要以林木为研究材料,开展林木遗传育种方面的研究。主持福建省自然科学基金面上项目一项,福建省协同创新项目一项。目前研究方向:1.以毛竹和杉木为材料,建立高效的组织快繁及遗传转化体系;2.利用基因组研究所取得的遗传信息,开展毛竹的重要性状(开花、速生与抗逆)的分子机制研究; 3. 植物基因工程研究,利用所建立的转基因技术,培育早开花、抗逆、及速生的竹类新品种;4. 濒危珍稀树种的扩繁技术研究。
邮箱(Email): zhuqiang@fafu.edu.cn
研究领域 (Research focus):
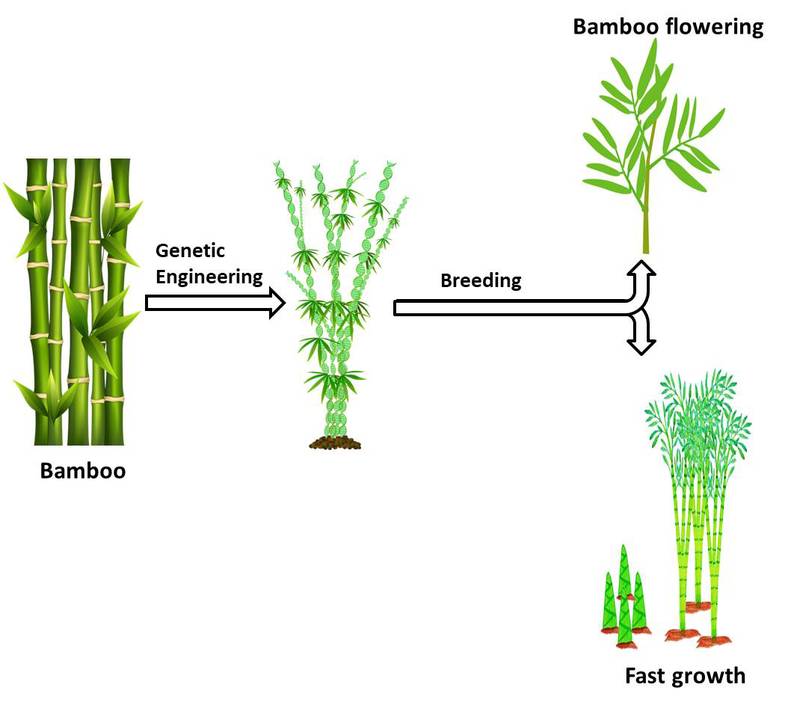
以毛竹和杉木为研究对象,开展林木遗传转化相关研究,研发林木遗传改良新体系,建立毛竹、杉木组织培养、快繁及遗传转化体系,以期培育早花、抗逆、速生的林木新品种,缩短林木生产周期,服务林业生产。
Our research mainly focuses on the genetic transformation of woody plants; we plan to establish the high efficiency platform for tissue culture and genetic transformation of Chinese fir and Maso bamboo; our goal is to construct woody plants with early flowering, stress tolerance and fast growth, which can reduce wood lifecycle and increase wood production.
发表文章(Publications):
2025
Wang W,Wu Q,Wang N,Ye S,Wang Y,Zhang J,Lin C,Zhu Q(2025) Advances in bamboo genomics: Growth and development, stress tolerance, and genetic engineering.J Integr Plant Biol.67(7):1725-1755.
Wang N,Wang W,Zhu Q.(2025)Unlocking the genetic blueprint of bamboo for climate adaption.Trends Plant Sci.30(7):693-695.
Liang J,Zhou L,Hu X,Lu J,Wang W,Zhu Q.(2025) Functional and expression profiling of DREB genes in Ma Bamboo (Dendrocalamus latiflorus Munro) reveals their role in abiotic stress adaptation.Plant Physiol Biochem.doi: 10.1016/j.
Cai C,Ye S,Liu J,Wang N,Tu M,Ding W,Wang W,Hu X,Wang Y,Ma X,Zhu Q.(2025) Functional insights into 4-coumarate-CoA ligase and caffeic acid O-methyltransferase genes involved in lignin biosynthesis in Chinese fir (Cunninghamia lanceolata).Int J Biol Macromol. doi: 10.1016/j.
Ding W,Ye S,Wu D,Wang W,Xu J,Wang W,Cai C,Lin C,Ma X,Zhu Q.(2025) Time-course transcriptome and proteomic dynamics during the de novo shoot organogenesis in Chinese fir (Cunninghamia lanceolata).Plant J. Aug;123(3):e70360.doi: 10.1111/tpj.70360.
2024
Wang W, Li Y, Cai C and Zhu Q.(2024) Auxin response factors fine-tune lignin biosynthesis in response to mechanical bending in bamboo. New Phytol.241:1161-1176.
2023
Hu X, Liang J, Wang W, Cai C, Ye S, Wang N, Han F, Wu Y and Zhu,Q.(2023) Comprehensive genome-wide analysis of the DREB gene family in Moso bamboo (Phyllostachys edulis): evidence for the role of PeDREB28 in plant abiotic stress response. Plant J.116: 1248-1270.
Wang N, Wang W, Cheng Y, Changyang C and Zhu Q.(2023) Uncovering the miRNA-mediated regulatory network involved in Ma bamboo (Dendrocalamus latiflorus) de novo shoot organogenesis. Horticulture research.10: uhad223.
Ye S, Ding W, Bai W, Lu J, Zhou L, Ma X and Zhu Q.(2023) Application of a novel strong promoter from Chinese fir (Cunninghamia lanceolate) in the CRISPR/Cas mediated genome editing of its protoplasts and transgenesis of rice and poplar. Frontiers in plant science.14.
2022
Wang K, Liu M, Cai C, Cai S, Ma X, Lin C and Zhu Q.(2022) The impact of genetic modified Ma bamboo on soil microbiome. Frontiers in Microbiology.13.
2021
Xiang M,Ding W,Wu C,Wang W,Ye S,Cai C,Hu X,Wang N,Bai W,Tang X,Zhu C,Xu Q,Zheng Y,Ding Z,Lin C,Zhu Q. (2021) Production of purple Ma bamboo (Dendrocalamus latiflorus Munro) with enhanced drought and cold stress tolerance by engineering anthocyanin biosynthesis. Planta. 254(3):50. doi: 10.1007/s00425-021-03696-z.
2020
Cai C, Wang W, Ye S, Zhang Z, Ding W, Xiang M, Wu C, Zhu Q. (2020) Overexpression of a Novel Arabidopsis Gene SUPA Leads to Various Morphological and Abiotic Stress Tolerance Alternations in Arabidopsis and Poplar. Front Plant Sci. doi: 10.3389/fpls.2020.560985.
2018
Zhang H, Wang H, Zhu Q, Gao Y, Wang H, Zhao L, Wang Y, Xi F, Wang W, Yang Y, Lin C, Gu L.(2018) Transcriptome characterization of moso bamboo (Phyllostachys edulis) seedlings in response to exogenous gibberellin applications BMC plant biology18(1):125. doi: 10.1186/s12870-018-1336-z.
2017
Ye S ,Cai C, Ren H, Wang W, Xiang M, Tang X, Zhu C, Yin T, Zhang L and Zhu Q.(2017) An Efficient Plant Regeneration and Transformation System of Ma Bamboo (Dendrocalamus latiflorus Munro) Started from Young Shoot as ExplantFront Plant Science8:1298 doi:10.3389/fpls.2017.01298.
Wang W,Gu L,Ye S,Zhang H,Cai C,Xiang M,Gao Y,Wang Q,Lin C and Zhu Q. (2017) Genome-wide analysis and transcriptomic profiling of the auxin biosynthesis,transport and signaling family genes in moso bamboo (Phyllostachys heterocycla)BMC Genomics 18:870 DOI 10.1186/s12864-017-4250-0.
Andrés-Colás N, Zhu Q, Takenaka M , Rybel B , Weijers D and Straeten D.(2017) Multiple PPR protein interactions are involved in the RNA editing system in Arabidopsis mitochondria and plastids PNAS 114(13):8883-8888.
Zhu Q, Zadnikova P, Smet D, Van Der Straeten D, Benkova E.(2017) Real-Time Analysis of the Apical Hook Development Methods in Molecular Biology1497:1-8.
2016
Zhu Q, Benková E.(2016) Seedlings' Strategy to Overcome a Soil Barrier Trends Plant Sciencepii: S1360-1385(16)30113-3.
Li WL, Liu Y, Douglas CJ. (2016) Role of glycosyl transferases in pollen wall primexine formation and exine patterning in Arabidopsis Plant PhysiologyDOI:10.1104/pp.16.00471.
2015
Liu Y, Douglas CJ. (2015) A role for OVATE FAMILY PROTEIN1 (OFP1) and OFP4 in a BLH6-KNAT7 multi-protein complex regulating secondary cell wall formation in Arabidopsis thalianaPlant Signaling& Behavior10(7): e1033126.
2014
Zhu, Q.,Dugardeyn, J., Zhang, C., Muhlenbock, P., Eastmond, P.J., Valcke, R., De Coninck, B., Oden, S., Karampelias, M., Cammue, B.P., Prinsen, E., and Van Der Straeten, D. (2014) The Arabidopsis petatricopepitide protein SLO2 participates in biotic and abiotic stress response Molecular Plant7 (2): 290-310.
Liu Y, You S, Taylor-Teeples M, Brady SM, Li L, Schuetz M, Douglas CJ.(2014) BEL1-LIKE HOMEODOMAIN6 (BLH6) and KNOTTED ARABIDOPSIS THALIANA7(KNAT7) interact and regulate secondary cell wall formation via repression of REVOLUTA Plant Cell 26:4843-4861.
Before 2014
Zhu, Q., E, H.M., and Van Der Straeten, D. (2012) SLO2, a mitochondrial PPR protein affecting several RNA editing sites, is required for energy metabolism The Plant Journal71:836-849.
Zhu, Q.,Dugardeyn, J., Zhang, C., Takenaka, M., Kuhn, K., Craddock, C., Smalle, J., Karampelias, M., Denecke, J., Peters, J., Gerats, T., Brennicke, A., Eastmond, P., Meyer, E.H., and Van Der Straeten, D. (2012) Functional analysis of SLO2 provides new insight into the role of plant PPR proteins Plant signaling & behavior7:1-3.
Zhang, J., Liu, H., Sun, J., Li, B., Zhu, Q., Chen, S. and Zhang, H. (2012) Arabidopsis Fatty Acid Desaturase FAD2 Is Required for Salt Tolerance during Seed Germination and Early Seedling Growth PLoS One7:e30355.
Zhu, Q., Zhang, J., Gao, X., Tong, J., Xiao, L., Li, W., and Zhang, H. (2010) The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses Gene457:1-12.
Zhang, J.T., Zhu, J.Q., Zhu, Q., Liu, H., Gao, X.S., and Zhang, H.X. (2009) Fatty acid desaturase6 (Fad6) is required for salt tolerance in Arabidopsis thaliana Biochem Biophys Res Commun390:469-474.











